Isolated molecule calculation protocol with Materials Studio
Comparison between calculated NMR parameters for an isolated molecule in a vacuum relative to a full crystal calculation offer insight into the intermolecular interactions, such as intermolecular hydrogen bonding and aromatic π-π stacking.
- Take a structure that has been geometry optimized using the specifications detailed within the protocol.
- Unbuild the crystal, which can be done under: Build > Symmetry > Unbuild Crystal.
- Afterwards the crystal should be rebuilt using: Build > Crystals > Rebuild Crystals, however the lattice parameters should be changed and a different symmetry operation used.
- Enter a ‘1 P1’ symmetry group, as shown in Fig. 1
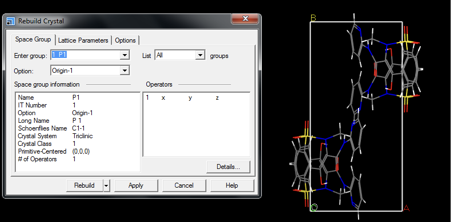
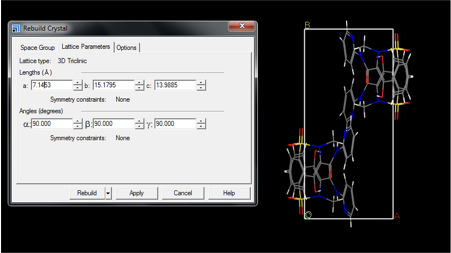
The lattice parameters should be chosen so that approximately 5 Å vacuum is present between molecules (see below for advice on how to achieve this). The effect of the neighboring molecule reduces with distance, although a larger box will increase the computational time. Furthermore, an orthorhombic cell is recommended. Lattice parameters are controlled in the Lattice Parameters tab (see Fig. 2).
To improve visualization of the molecule in the box, and to ensure that the box is of a sufficient size, it is recommended that the molecule is translated to the centre of the box. This step is solely for visualisation and comparison reasons only, and has no effect on the calculation. To move the molecule select all atoms using Edit > Atom Selection > Select all. The molecule (or all atoms) can be moved in (x,y,z) using move left, move right, move up and move down buttons, as shown in Fig. 3.
For Z’ > 1, separate calculations are required for each molecule in the asymmetric unit cell.
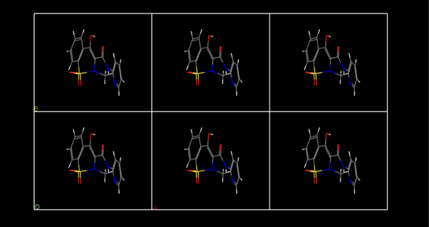
Once the molecule is centred in a box in all dimensions, the lattice parameters should be changed using the same procedure shown in Fig. 2 so that the box is approximately the same size as the molecule (within 1 Å), as shown for piroxicam in Fig. 4a. This may require further adjustment using the buttons shown in Fig. 3. Once this is achieved, lattice parameters a, b, and c should each be increased by 5 Å. Fig. 4b shows the actual periodicity present and that the calculation is for an isolated, as opposed to single, molecule and still exploits the periodicity of a crystalline material.
![Figure 4a: Geometry-optimized β-piroxicam structure [BIYSEH03] as a single molecule in a box. The box shown approximately matches the molecule dimensions. For an isolated molecule calculation, 5 Å should then be added to each lattice parameter a, b, c.](/user/pages/05.docs/05.materials-studio-isolated/iso_fig4a.png)
- A NMR calculation is performed in the same manner as outlined within the Materials Studio CASTEP protocol. Ensure to select Energy dropdown in Task box and tick NMR in properties tab.
- The unit cell must be fixed. Consequently, a dispersion scheme is no longer strictly necessary (although it is not critical if included).
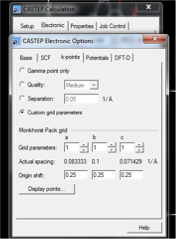
- The cut-off energy used should match full crystal calculations, herein 700 eV is recommended. For a molecule in a super vacuum single cell it is appropriate to sample with a single k-point. The most efficient k-point spacing for an isolated molecule is therefore using a single k-point situated at (¼, ¼, ¼). This is achieved with the Electronic tab, click More... and specify Custom grid parameters. a, b, and c set to 1, 1, 1, and the origin shift should be set to 0.25, 0.25, 0.25 (see Fig. 5). Using this point gives a more realistic approximation of the wave function in the 1st Brillouin Zone relative to the Γ-point, and reduces the size of the box required to achieve convergence of the calculated shielding values, as shown in Fig. 6.
![Figure 6: Convergence of calculated absolute shielding values of an isolated molecule of methane with respect to box size using a k-point offset of [0,0,0] [black line] and a k-point offset of [¼, ¼, ¼] [red line]. Reproduced with permission from Prof. Jonathan Yates.](/user/pages/05.docs/05.materials-studio-isolated/iso_fig6.png)
The NMR calculations are performed in the same manner as detailed in the methodology outlined within the CASTEP NMR using Materials Studio document.
Acknowledgements
The protocol described here is a product of a collaborative effort of the following:
- Andrew S. Tatton and Jonathan R. Yates, Department of Materials, Oxford University, Oxford, UK
- Les Hughes and Helen Blade, Pharmaceutical Development, AstraZeneca, Macclesfield, UK
- Sten Nilsson Lill, Early Product Development, AstraZeneca, Mölndal, Sweden
- Albert Bartók-Pártay, STFC Scientific Computing Department, Rutherford Laboratory, Didcot, UK
- Steven P. Brown, Department of Physics, University of Warwick, Coventry, UK
- Paul Hodgkinson, Department of Chemistry, Durham University, Durham, UK
Andrew S. Tatton performed the calculations and prepared this document.